Microbial communities: The bad guys, the good guys, and… the antibiotic pressure
By: Ximena Serrano Gil
Photos: 123RF, Milagro Castro
Health and Wellness
By: Ximena Serrano Gil
Photos: 123RF, Milagro Castro
Infections associated with intestinal bacteria are a major problem worldwide, as they favor the onset of life-threatening diseases in humans and animals. From the Universidad del Rosario, Marina Muñoz Díaz, PhD in Biotechnology, leads studies aimed at the detection and characterization of pathogenic bacteria, which affect the wellbeing of an individual and can cause changes in the microbial community. Dr. Muñoz’s work, together with researchers from the Microbiology and Biotechnology Research Center of the Universidad del Rosario (Cimbiur) of the Faculty of Natural Sciences, of which she is the technical director, is pioneering in Latin America.
Identifying the diversity of species that colonize an organism (microbiota) and that compete, cooperate, and interact with each other is essential to ensure the effectiveness of existing treatments against them.
In this regard, the researcher explains that “what is transversal in microbiology research is the use of molecular biology and next-generation sequencing tools, which make it possible to understand the genetic bases that modulate the effect of microbial species (bacteria, fungi, parasites, and viruses) on hosts and environments. This interaction produces both beneficial and detrimental effects on human, animal, and environmental health.”
“My research is initially aimed at characterizing the ‘bad guys’, i.e., the pathogens. Once we know the genetic bases, we will be able to find solutions to serious health problems,” says Muñoz.
This characterization is unique for each patient and allows the development of a map that serves as a reference. Nevertheless, the description has to be periodically repeated since microorganisms change, i.e., they gain and lose genetic material.
For example, “we can have Escherichia coli, a common bacterium in the intestinal microbiota. Some populations of this species are bad (pathogenic) and, if they have the opportunity to multiply, can cause serious problems. If, in addition, they are resistant to antibiotics, a treatment that was once effective now becomes ineffective. Therefore, the patient’s clinical condition may become more complicated until a truly effective alternative is found. This is why characterizing the ‘bad guys’ is so important,” adds the researcher.
The tools used in Cimbiur allow the identification of the presence of microorganisms and molecular markers of interest for health, such as those related to antibiotic resistance, which provides very useful information for patient management.
The researcher’s main work focuses on the characterization of bacteria such as Clostridiodes difficile (which causes diarrhea and colitis), Clostridium perfringens (common in food poisoning), carbapenemase- producing enterobacteriaceae (CPE), and other microorganisms that severely affect the intra- hospital environment and the community.
On the other hand, “we hardly ever pay attention to the beneficial aspect, let alone consider a holistic view of the intestinal ecosystem, an essential approach to assessing the effects that occur. The analysis of the presence and interactions of pathogenic microorganisms, along with all the genetic material of the microbiota (the microbiome), is very important to learn about microbial ecology,” explains Muñoz.
“It is important to put all this information to use. Therefore, at Cimbiur, we plan to obtain products with biotechnological potential, such as diagnostic tests, surveillance schemes, and even probiotics, to transfer knowledge. Notably, science can have a positive impact on people by generating solutions to community problems,” says Professor Muñoz.
Antibiotic resistance is one of the leading causes of death worldwide. More than 1.2 million people died in 2019 as a direct result of infections with antibiotic-resistant bacteria, according to a study conducted in 204 countries and published in the journal The Lancet.
Although antibiotics are designed to cure and save lives, they can also cause adverse effects by eliminating beneficial intestinal bacteria responsible for maintaining the balance of the gastrointestinal system. In addition, antibiotics can exert a selection pressure that causes resistant populations, those with molecular markers conferring resistance, to proliferate. In such a situation, bacteria proliferate and invade the organism.

“My research is initially aimed at characterizing the ‘bad guys’, i.e., the pathogens. Once we know the genetic bases underlying their effect, we will be able to find solutions to serious health problems,” says Marina Muñoz Díaz, PhD in Biotechnology, who leads several studies at the Universidad del Rosario aimed at the detection and characterization of pathogenic bacteria.
The scientific community is warning that bacterial resistance to antibiotics can be the next pandemic. In the article Gut microbiota composition in health-care facility-and community-onset diarrheic patients with Clostridioides difficile infection, published in 2021 in the journal Scientific Reports of the publisher Nature, Professors Marina Muñoz and Juan David Ramírez stated that “one of the infections with the greatest global impact is Clostridioides difficile infection (CDI), considered to be the causative agent of diarrhea associated with the use of antibiotics.”
This bacterium can trigger a variety of mild problems, such as diarrhea and inflammatory bowel disease, to serious digestive disorders, such as toxic megacolon, pseudomembranous colitis, and sepsis, which entail a high mortality risk. The problems associated with CDI worsen mainly in patients in intensive care units (ICUs), where it represents one of the five infections with the greatest impact worldwide.
These disorders are related to the overuse or misuse of antibiotics, which exposes the organism to a stimulus that affects the microbial communities. Consequently, bacteria capable of resisting antibiotics proliferate and colonize the intestinal micro environment. According to Muñoz, “we are attacking our organism in two ways, i.e., attacking the beneficial microbiota and favoring the pathogen. In short, we are disrupting the intestinal balance.”
Patients in ICUs are administered high doses of antibiotics that cause an imbalance in the microbial communities, which, together with immunosuppression, either due to existing diseases or to the application of treatments, makes these patients more susceptible to suffer complications such as CDI. “It is the ideal environment for pathogenic bacteria to multiply, colonize, and impair the balance of the intestinal microbiota. This is associated, among other factors, with poor absorption of nutrients and weakening of the immune system,” explains Muñoz. Therefore, this population is of great interest to the research center, whose members are also involved in a collaborative project with the Hospital Universitario Mayor Méderi in Bogotá, aimed at describing the intestinal metagenome in ICU patients.
The study published in Scientific Reports described the changes observed in microbial communities in patients with C. difficile. The hospital is now studying all the genetic material, known as the metagenome, to fully understand what is happening to patients at the intrahospital level.
According to biologist Muñoz, previous studies have shown that there is a high frequency of infection in both intrahospital and community settings. Using molecular techniques, the microorganism (C. difficile) has been detected in more than 58 percent of hospitalized individuals and in approximately 38 percent of people in the community. Depending on individual risk factors, complex clinical outcomes can occur.
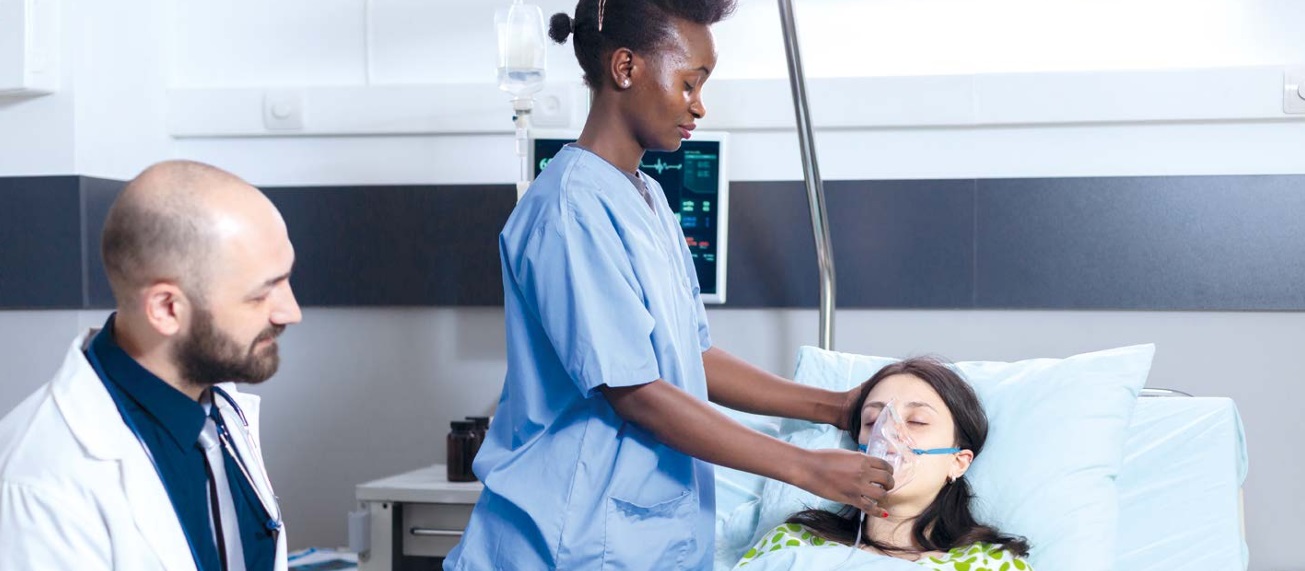
Patients in ICUs are administered high doses of antibiotics that cause an imbalance in the microbial communities, which, together with immunosuppression, either due to existing diseases or to the application of treatments, makes these patients more susceptible to suffer complications.
The microbial diversity of the country has not been thoroughly studied. In general, the diversity of plants and animals is discussed but not that of microorganisms, which are even more diverse. “Our research center is working on the description of microbial diversity and reporting it in international databases because we need the world to know it to develop more accurate comparison strategies. This research is pioneering in Colombia,” says Muñoz.
According to the biologist, research on C. difficile carried out in the United States and England has shown that the populations of microorganisms are highly conserved, i.e., they are similar to each other; diversity has been limited to a few clades (a group of species that share a common ancestor). By contrast, studies in Colombia found that there is a high diversity and that there are more populations of these microorganisms than previously reported.
Upon studying the molecular markers (small windows within the genome) and observing that there were different arrangements, the researchers decided to analyze the entire genome. The research center is also a pioneer in the “study of genomic epidemiology,” which makes it possible to gain insight into the organization of the human genome and, at the same time, to carry out robust studies on populations of microorganisms.
The next step of the research consisted of studying whether the microbial population in hospitalized patients was the same as that found in the community (outside the hospital). Using the tool, it was determined that only one of these populations was shared.
The microbiomes inside and outside the ICUs function with a different structure at the genome level, including the genes that encode toxins and the antibiotic resistance markers. Taken together, this may increase the complexity of the clinical management of infected patients, which has not been previously studied in Colombia.
“If physicians had access to the results obtained from this type of tools for each patient, they could easily select and prescribe the most effective treatment. These tools are also useful to know the dispersion patterns of these microorganisms and to discover the way they reach different regions or even the way they spread within a hospital,” emphasizes Muñoz.
Clostridioides difficile is a grampositive, anaerobic bacterium that forms resistant spores, which can be found in contaminated food or water. The microorganism reaches the intestine through the consumption of spores, where it produces an array of toxins that cause the associated diseases.
The spores are shed through the feces and are very resistant, even to sodium hypochlorite. Therefore, it is advisable to thoroughly decontaminate vegetables by hand.
Infection levels Asymptomatic patients. Patients with diarrhea. Simple colitis.
All of the above refers to the controllable part of the infection, but when other variables come into play, such as overuse of antibiotics, other conditions, or a patient who is immunocompromised, the impact of the microorganisms can be exacerbated and result in Pseudomembranous colitis. Fulminant colitis/toxic megacolon. Death of the patient.
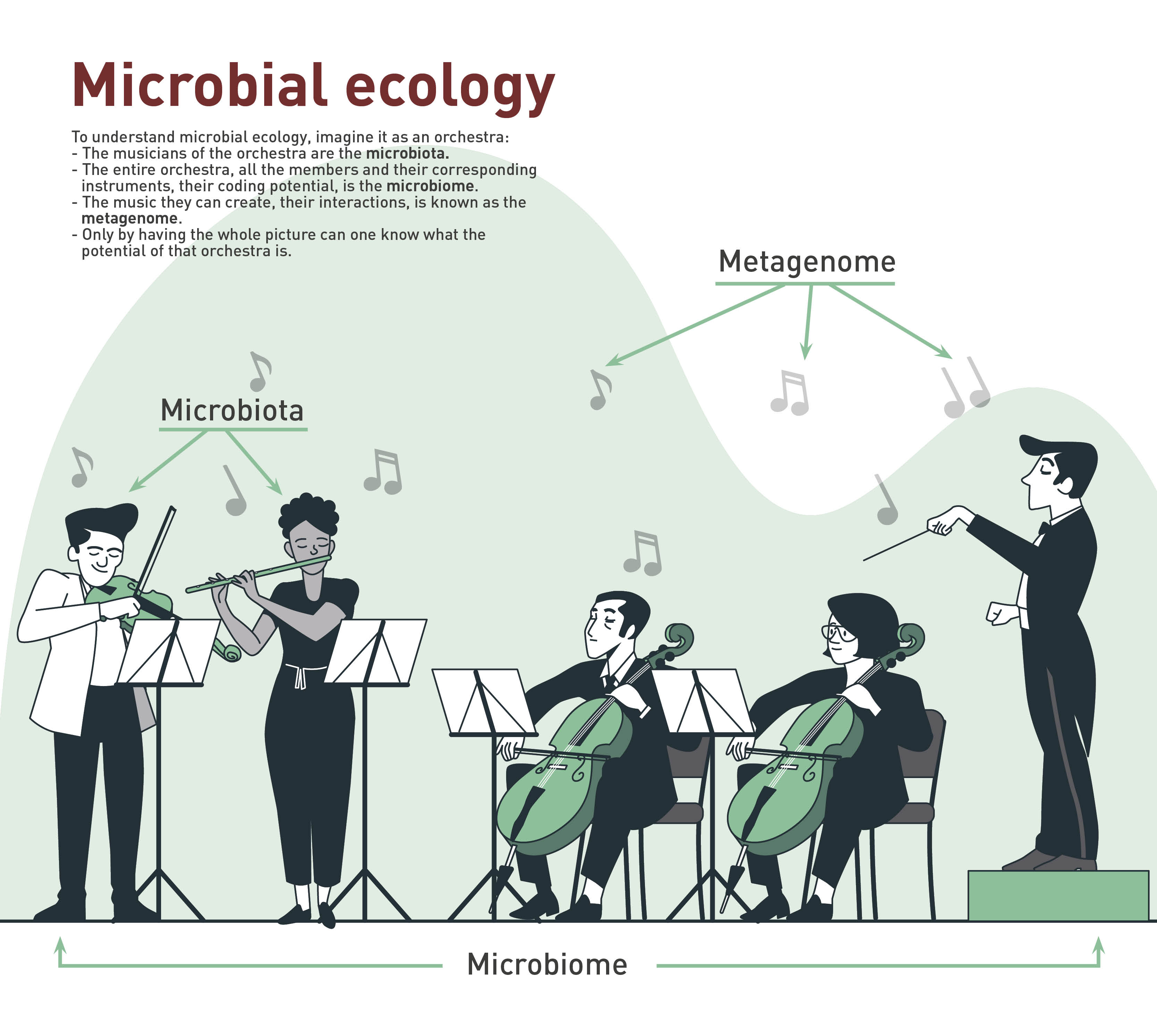
Microbiota: Group of microorganisms found in an environment (skin, mucous membranes, etc.). Microorganisms inhabit these places on a more or less permanent basis and, in some cases, have specific functions.
Microbiome: It refers to the microorganisms present in a specific environment, including all the genetic material. This group forms a dynamic and interactive micro ecosystem.
Metagenome: The set of microbial genes present in a given environment or ecosystem. Metagenomics includes the techniques used to study this whole set without the need to isolate each of the population found.